The Distribution of Cherry Trees in Washington DC
1 Introduction
I had long wished to see cherry blossom in Washington DC. However, this year has been a year of staying in, which makes me even more eager to see it. As a result, I pulled up cherry trees data from the open data portal and explored it before I can see it in person. In this notebook, I will first use ggplot2 to map the cherry data and then play with leaflet to try the interactive visualization.
2 Data Preparation
2.1 Set Up
2.2 Read in data and clean
## # A tibble: 6 x 39
## X Y OBJECTID FACILITYID VICINITY WARD TBOX_L TBOX_W WIRES CURB SIDEWALK TBOX_STAT
## <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr>
## 1 -77.0 38.9 43998460 10110-185~ 1810 11~ 1 9 4 None Perm~ Permane~ Plant
## 2 -77.0 38.9 43998509 <NA> 2200 BL~ 6 99 9 None None None Plant
## 3 -77.0 38.9 43998510 28801-200~ 2000 UP~ 5 99 3 Both Perm~ Permane~ Plant
## 4 -77.1 38.9 43998511 10393-230~ 2408 39~ 3 99 4 None Perm~ Permane~ Plant
## 5 -77.0 38.9 43998512 <NA> 2200 BL~ 6 99 9 None None None Plant
## 6 -77.0 38.9 43998513 36981-290~ 2927 PE~ 7 99 6 Both Perm~ Permane~ Plant
## # ... with 27 more variables: RETIREDDT <chr>, SCI_NM <chr>, CMMN_NM <chr>, DATE_PLANT <chr>,
## # DBH <dbl>, DISEASE <chr>, PESTS <chr>, CONDITION <chr>, CONDITIODT <chr>, OWNERSHIP <chr>,
## # TREE_NOTES <chr>, ONEYEARPHOTO <chr>, SPECIALPHOTO <lgl>, PHOTOREMARKS <lgl>,
## # ELEVATION <chr>, SIGN <chr>, TRRS <dbl>, WARRANTY <chr>, FAM_NAME <chr>, CREATED_USER <chr>,
## # CREATED_DATE <chr>, EDITEDBY <chr>, LAST_EDITED_USER <chr>, LAST_EDITED_DATE <chr>,
## # GENUS_NAME <chr>, GLOBALID <chr>, SHAPE <lgl>## variable q_zeros p_zeros q_na p_na q_inf p_inf type unique
## 1 X 0 0.00 2 0.00 0 0 numeric 190378
## 2 Y 0 0.00 2 0.00 0 0 numeric 190380
## 3 OBJECTID 0 0.00 0 0.00 0 0 numeric 190769
## 4 FACILITYID 3 0.00 38324 20.09 0 0 character 149091
## 5 VICINITY 0 0.00 3404 1.78 0 0 character 115842
## 6 WARD 0 0.00 766 0.40 0 0 numeric 8
## 7 TBOX_L 20 0.01 17801 9.33 0 0 numeric 99
## 8 TBOX_W 24 0.01 17763 9.31 0 0 numeric 81
## 9 WIRES 0 0.00 3467 1.82 0 0 character 4
## 10 CURB 0 0.00 3311 1.74 0 0 character 4
## 11 SIDEWALK 0 0.00 3453 1.81 0 0 character 4
## 12 TBOX_STAT 0 0.00 95 0.05 0 0 character 8
## 13 RETIREDDT 0 0.00 0 0.00 0 0 character 4305
## 14 SCI_NM 0 0.00 15951 8.36 0 0 character 206
## 15 CMMN_NM 0 0.00 14971 7.85 0 0 character 205
## 16 DATE_PLANT 0 0.00 0 0.00 0 0 character 43530
## 17 DBH 4714 2.47 15785 8.27 0 0 numeric 603
## 18 DISEASE 0 0.00 188871 99.01 0 0 character 11
## 19 PESTS 0 0.00 189673 99.43 0 0 character 5
## 20 CONDITION 0 0.00 14881 7.80 0 0 character 5
## 21 CONDITIODT 0 0.00 0 0.00 0 0 character 153216
## 22 OWNERSHIP 0 0.00 1135 0.59 0 0 character 7
## 23 TREE_NOTES 7 0.00 115204 60.39 0 0 character 34001
## 24 ONEYEARPHOTO 0 0.00 189991 99.59 0 0 character 28
## 25 SPECIALPHOTO 0 0.00 190768 100.00 0 0 logical 1
## 26 PHOTOREMARKS 0 0.00 190769 100.00 0 0 logical 0
## 27 ELEVATION 245 0.13 11773 6.17 0 0 character 7
## 28 SIGN 0 0.00 12744 6.68 0 0 character 6
## 29 TRRS 53 0.03 190431 99.82 0 0 numeric 11
## 30 WARRANTY 0 0.00 24317 12.75 0 0 character 17
## 31 FAM_NAME 0 0.00 20812 10.91 0 0 character 86
## 32 CREATED_USER 0 0.00 151684 79.51 0 0 character 38
## 33 CREATED_DATE 0 0.00 0 0.00 0 0 character 34899
## 34 EDITEDBY 0 0.00 5648 2.96 0 0 character 40
## 35 LAST_EDITED_USER 0 0.00 19 0.01 0 0 character 40
## 36 LAST_EDITED_DATE 0 0.00 0 0.00 0 0 character 152696
## 37 GENUS_NAME 0 0.00 25420 13.33 0 0 character 77
## 38 GLOBALID 0 0.00 0 0.00 0 0 character 190769
## 39 SHAPE 0 0.00 190769 100.00 0 0 logical 0Rename variables to meaningful names and filter out rows that lack longitude or latitude.
trees <- trees %>%
rename(long= X, lat=Y, plant_names=CMMN_NM) %>%
filter(!is.na(long) & !is.na(lat) & !is.na(plant_names))Filter trees that has “cherry” in the plant names.
cherry_trees <- trees %>%
filter(str_detect(trees$plant_names, regex(".*cherry.*", ignore_case=TRUE))) %>%
select(long, lat, plant_names)Now we have the cherry data, we would like to see the cultivars and the quantity of each cultivar.
## Warning: `...` is not empty.
##
## We detected these problematic arguments:
## * `needs_dots`
##
## These dots only exist to allow future extensions and should be empty.
## Did you misspecify an argument?## # A tibble: 7 x 2
## plant_names n
## <chr> <int>
## 1 Cherry 2209
## 2 Kwanzan cherry 2038
## 3 Chokecherry 1929
## 4 Yoshino cherry 1835
## 5 Cherry (Snowgoose) 1813
## 6 Okame cherry 855
## 7 Cherrybark Oak 5The most common cultivar is cherry without explicitly identifying its cultivar. There’s no other evidence here that would help us determine what it is, so we will just go with its original name- Cherry. In addition, after a bit of googling, it turns out that Cherrybark Oak is not a cherry tree, although it has cherry in its name. Thus I will exclude that from the data and in the meantime, update Chokecherry and Cherry (Snowgoose) to make all of the names have the same pattern.
cherry_trees <- cherry_trees %>%
filter(plant_names!="Cherrybark Oak") %>%
mutate(plant_names=case_when(.$plant_names=="Chokecherry" ~ "Choke cherry",
.$plant_names =="Cherry (Snowgoose)" ~ "Snowgoose cherry",
TRUE ~ .$plant_names
))Up till now the cherry tree data is ready. To map the cherry tree location to the map of DC, we need a DC map with neighborhoods boundaries.
2.3 DC shapefile
Read in the shapefile using rgdal package.
#create a folder to hold the dc shape files
dir.create("dc_shape1")
unzip("Neighborhood_Clusters-shp.zip", exdir = "dc_shape1")
#read in the dc shape file
shp <- readOGR("dc_shape1/Neighborhood_Clusters.shp")## OGR data source with driver: ESRI Shapefile
## Source: "C:\Users\wensh\Desktop\dccherrytrees\dc_shape1\Neighborhood_Clusters.shp", layer: "Neighborhood_Clusters"
## with 46 features
## It has 7 fieldsNow both of the cherry tree and map data are ready, it is time to plot them out and see the distribution of the cherry.
3 Data Visualization
3.1 ggplot2 Basic visualization
ggplot()+geom_polygon(data=shp, aes(long, lat, group=group), colour="black", fill=NA)+geom_point(data=cherry_trees, aes(long, lat))+theme_void()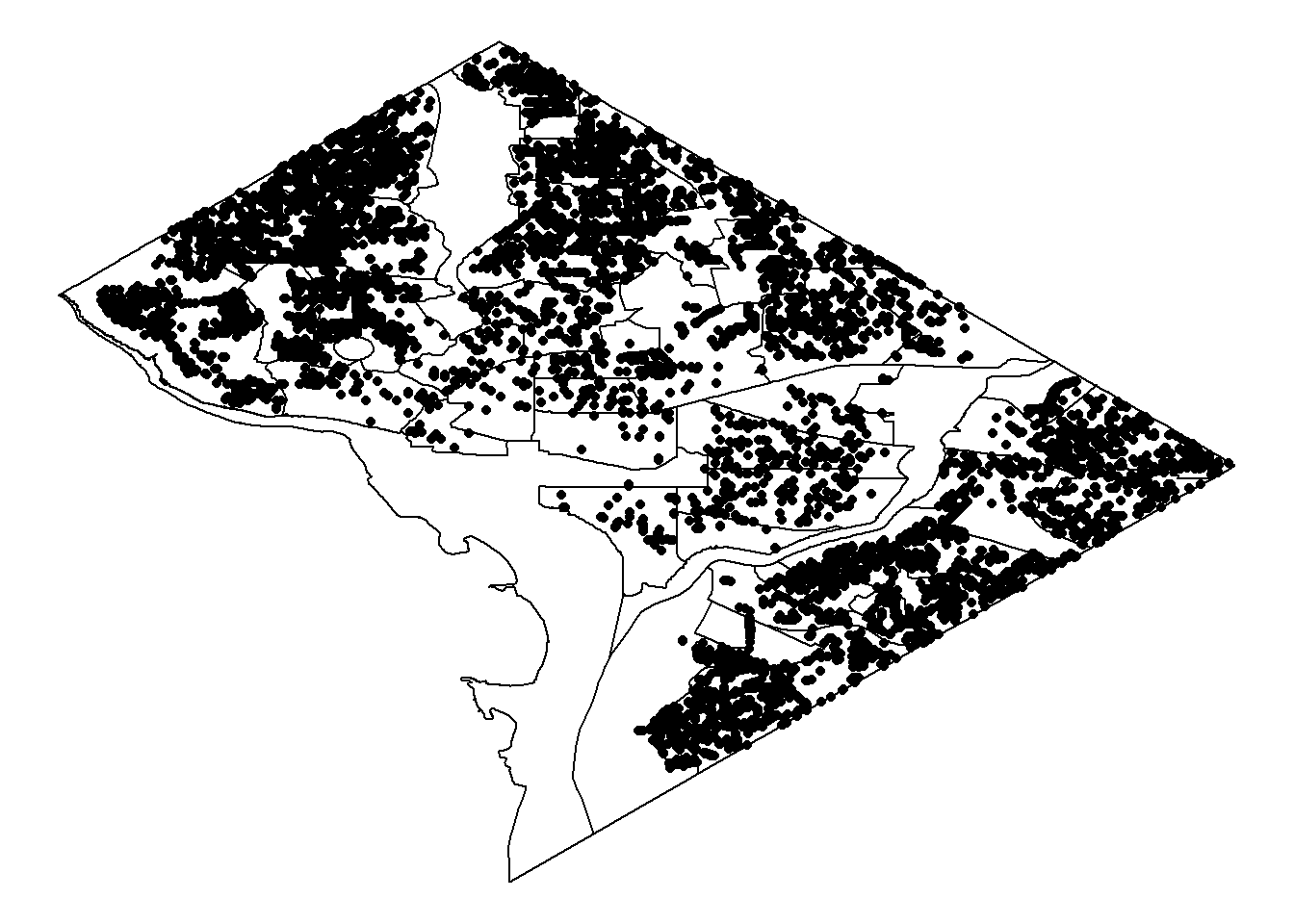
The basic visualization is hard to detect the number of trees in the communities. Hex geom can better plot the density. In the following, I will change the geom to hex and the colour scheme to fit in the cherry blossom atmosphere!
3.1.1 Geom and Color Change
p <- ggplot()+geom_polygon(data=shp, aes(long, lat, group=group), colour="black", fill=NA)+
geom_hex(data=cherry_trees, aes(long, lat))+
scale_fill_gradient(name="No. of trees", low="#ffcee6", high = "#6B0772")+theme_void()
print(p)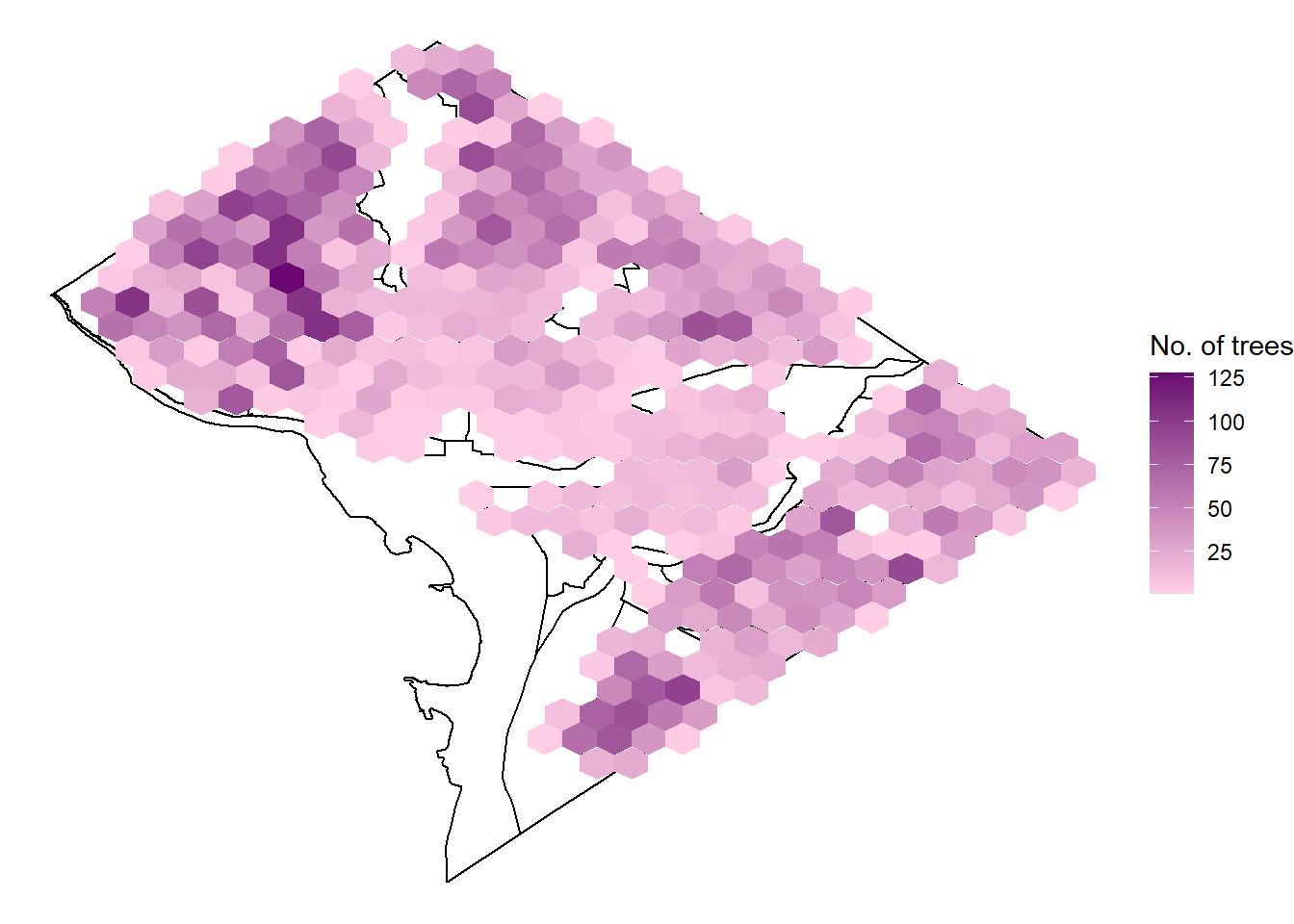
3.1.2 Distribution of Different Cultivars
Sometimes people have their preference when it comes to what kind of cherry trees to see, here I will subset the trees by facet_wrap() so people will know where to go based on the type of trees they prefer.
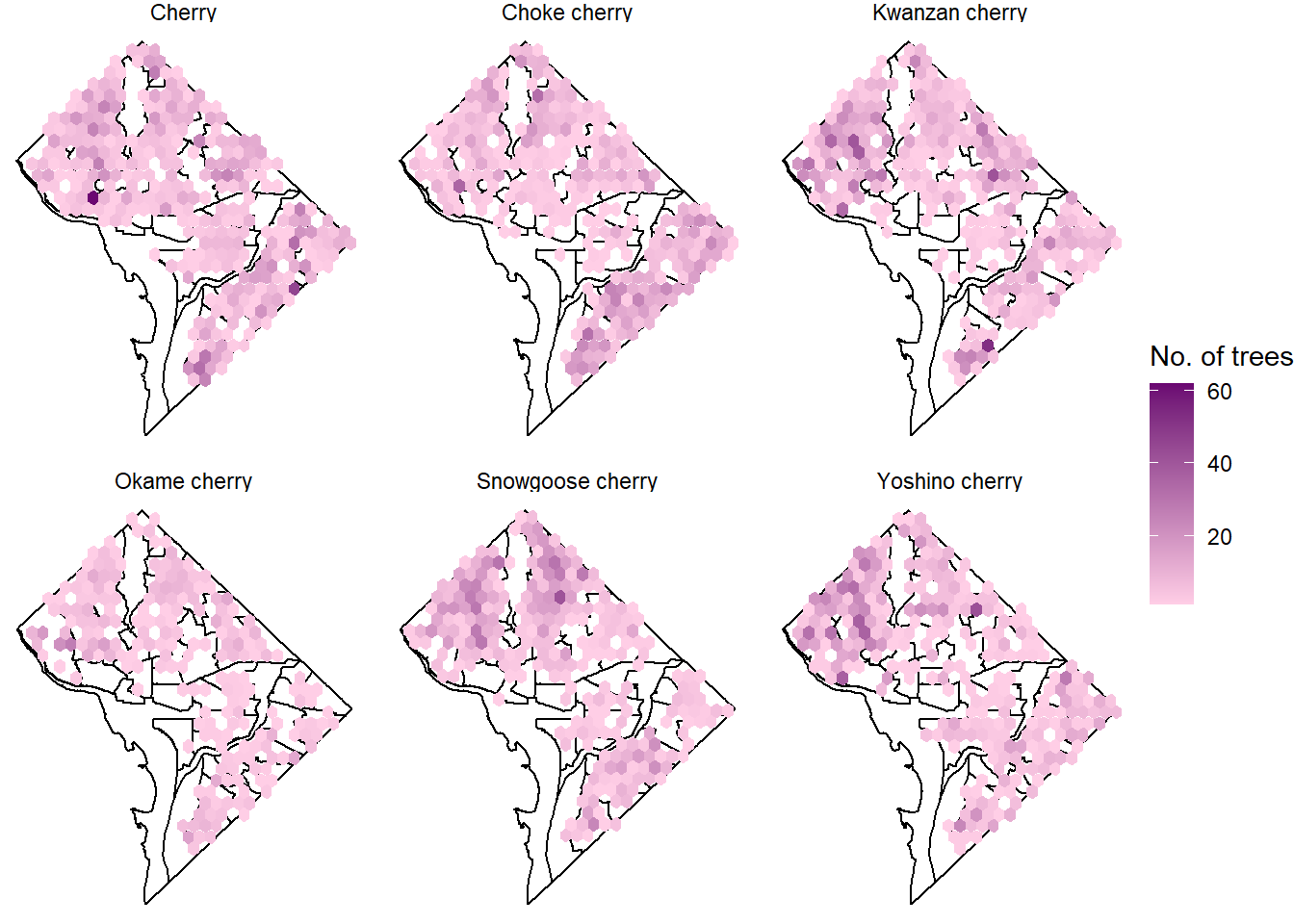
# customize the plots
p+labs(title=" The Distribution of Cherry Trees in Washington DC",
caption="data source: Open Data DC 2020")+
theme(plot.title=element_text(hjust = 0.5, margin=margin(b=16)),
legend.position = "bottom", legend.direction = "horizontal",
legend.title = element_text(vjust=0.9, size = 8),
legend.text = element_text(size=8),
legend.box.spacing = unit(1, "cm"),
legend.key.size = unit(4, "mm"),
plot.caption = element_text(face = "italic", color = "#696969"))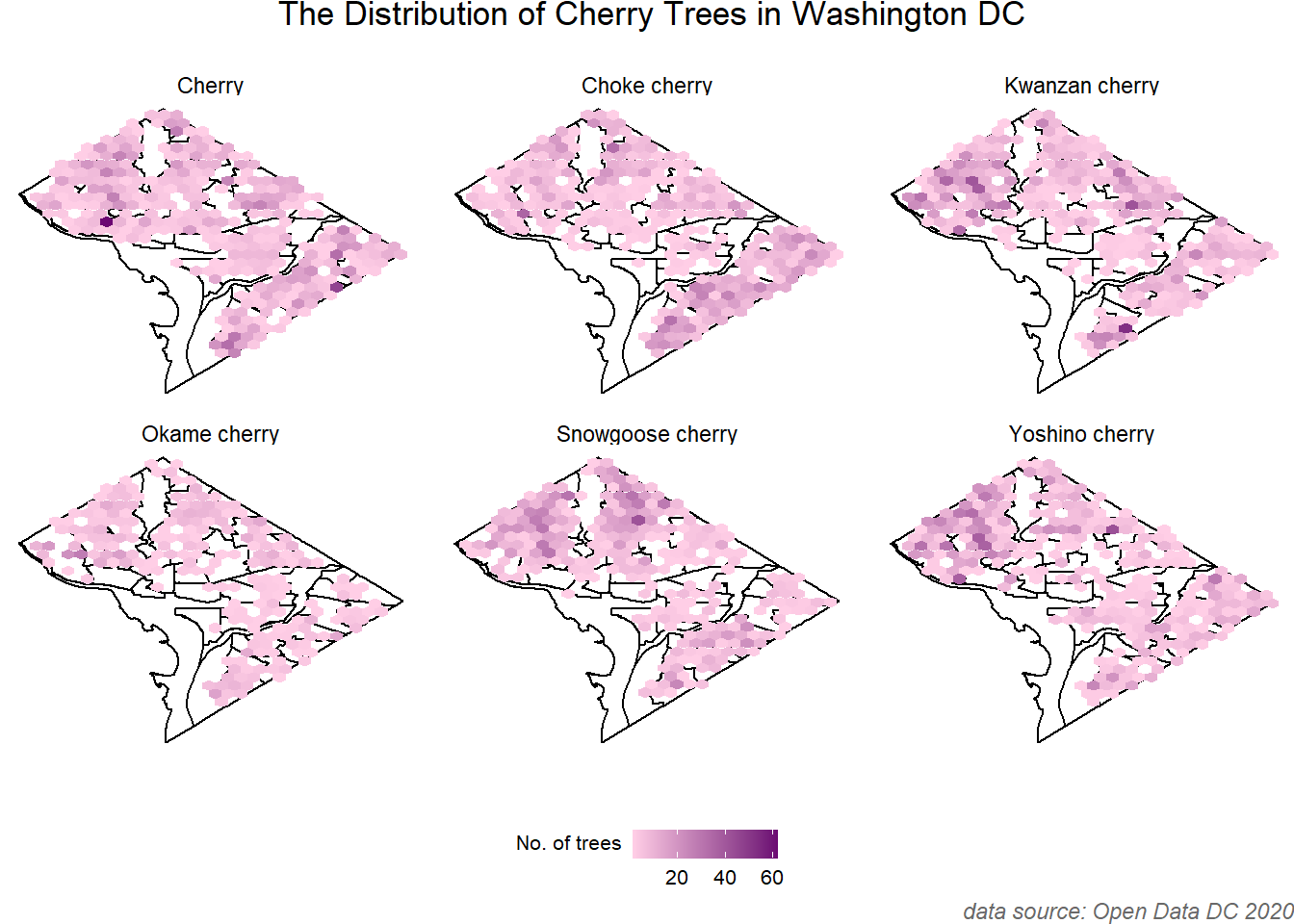
I tried to add the neighborhoods names to the plots but it made the plots a mess. Next I will try to utilize leaflet package to build an interactive map with the neighborhoods names shown on the map.
3.2 Interactive visualization
3.2.1 DC Interactive map with neighborhood names shown
First, I will use DC shape data shp to create an interactive map where you can see the corresponding neighborhood names when hovering over.
3.2.2 Cherry Data Added to the Map
Next, cherry data will be added as another layer to the map.
leaflet() %>%
addTiles() %>%
addPolygons(data=shp,
label = ~NBH_NAMES,
color = "444444",
weight=1,
smoothFactor = 0.5,
opacity = 1.0,
fillOpacity = 0.5,
highlightOptions = highlightOptions(color = "white",
weight = 2,
bringToFront = TRUE)) %>%
addMarkers(data=cherry_trees, clusterOptions = markerClusterOptions())A few places we can tell from the map are good for appreciation of cherry blossom: Twining, Cleveland Park, Brightwood Park and Congress Heights.
3.2.3 Seprate Maps for Different Cultivars
For people who have specific interests in certain kinds of cherry trees, below are the plots subsetting from the main one based on the type of cherry trees.
Cherry
leaflet() %>%
addTiles() %>%
addPolygons(data=shp,
label = ~NBH_NAMES,
color = "444444",
weight=1,
smoothFactor = 0.5,
opacity = 1.0,
fillOpacity = 0.5,
highlightOptions = highlightOptions(color = "white",
weight = 2,
bringToFront = TRUE)) %>%
addMarkers(data=cherry_trees %>% filter(plant_names=="Cherry"),
clusterOptions = markerClusterOptions())Places like Twinging, Brightwood Park and Brookland are densely distributed with Cherry.
Yoshino cherry
leaflet() %>%
addTiles() %>%
addPolygons(data=shp,
label = ~NBH_NAMES,
color = "444444",
weight=1,
smoothFactor = 0.5,
opacity = 1.0,
fillOpacity = 0.5,
highlightOptions = highlightOptions(color = "white",
weight = 2,
bringToFront = TRUE)) %>%
addMarkers(data=cherry_trees %>% filter(plant_names=="Yoshino cherry"),
clusterOptions = markerClusterOptions())Clearly, Yoshino cherry trees are more densely grown in the northwest of the city. More specifically, Friendship Heights is the place with most Yoshino.
Choke cherry
leaflet() %>%
addTiles() %>%
addPolygons(data=shp,
label = ~NBH_NAMES,
color = "444444",
weight=1,
smoothFactor = 0.5,
opacity = 1.0,
fillOpacity = 0.5,
highlightOptions = highlightOptions(color = "white",
weight = 2,
bringToFront = TRUE)) %>%
addMarkers(data=cherry_trees %>% filter(plant_names=="Choke cherry"),
clusterOptions = markerClusterOptions())People who have an penchant for Choke cherry should go to the east and southeast of DC. Places like Sheridan and Deanwood are good for Choke cherry.
Kwanzan cherry
leaflet() %>%
addTiles() %>%
addPolygons(data=shp,
label = ~NBH_NAMES,
color = "444444",
weight=1,
smoothFactor = 0.5,
opacity = 1.0,
fillOpacity = 0.5,
highlightOptions = highlightOptions(color = "white",
weight = 2,
bringToFront = TRUE)) %>%
addMarkers(data=cherry_trees %>% filter(plant_names=="Kwanzan cherry"),
clusterOptions = markerClusterOptions())Most of the Kwanzan cherry are located in the west of DC. If people go to Spring Valley and Friendship Heights, they should have a blast at the Kwanzan cherry.
Snowgoose cherry
leaflet() %>%
addTiles() %>%
addPolygons(data=shp,
label = ~NBH_NAMES,
color = "444444",
weight=1,
smoothFactor = 0.5,
opacity = 1.0,
fillOpacity = 0.5,
highlightOptions = highlightOptions(color = "white",
weight = 2,
bringToFront = TRUE)) %>%
addMarkers(data=cherry_trees %>% filter(plant_names=="Snowgoose cherry"),
clusterOptions = markerClusterOptions())If you are a big fan of Snowgoose cherry, you know where you should go- Cleveland Park, Friendship Heights, Brightwood Park and Takoma!
Okame cherry
leaflet() %>%
addTiles() %>%
addPolygons(data=shp,
label = ~NBH_NAMES,
color = "444444",
weight=1,
smoothFactor = 0.5,
opacity = 1.0,
fillOpacity = 0.5,
highlightOptions = highlightOptions(color = "white",
weight = 2,
bringToFront = TRUE)) %>%
addMarkers(data=cherry_trees %>% filter(plant_names=="Okame cherry"),
clusterOptions = markerClusterOptions())The west side of DC boasts the most Okame cherry trees. Spring Valley, Friendship Heights and Takoma are great to see Okame.
When the next cherry blossom season sets in, I hope you have an idea where you want to go for the best experience!